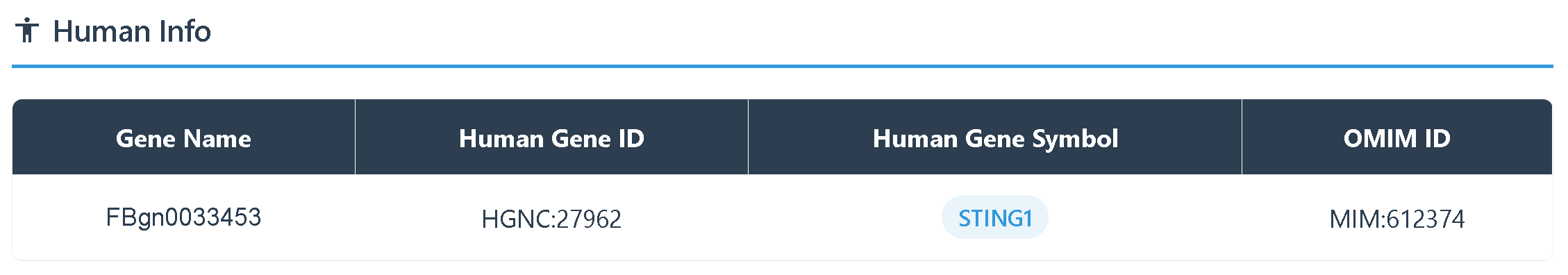
Overview of DrosGB
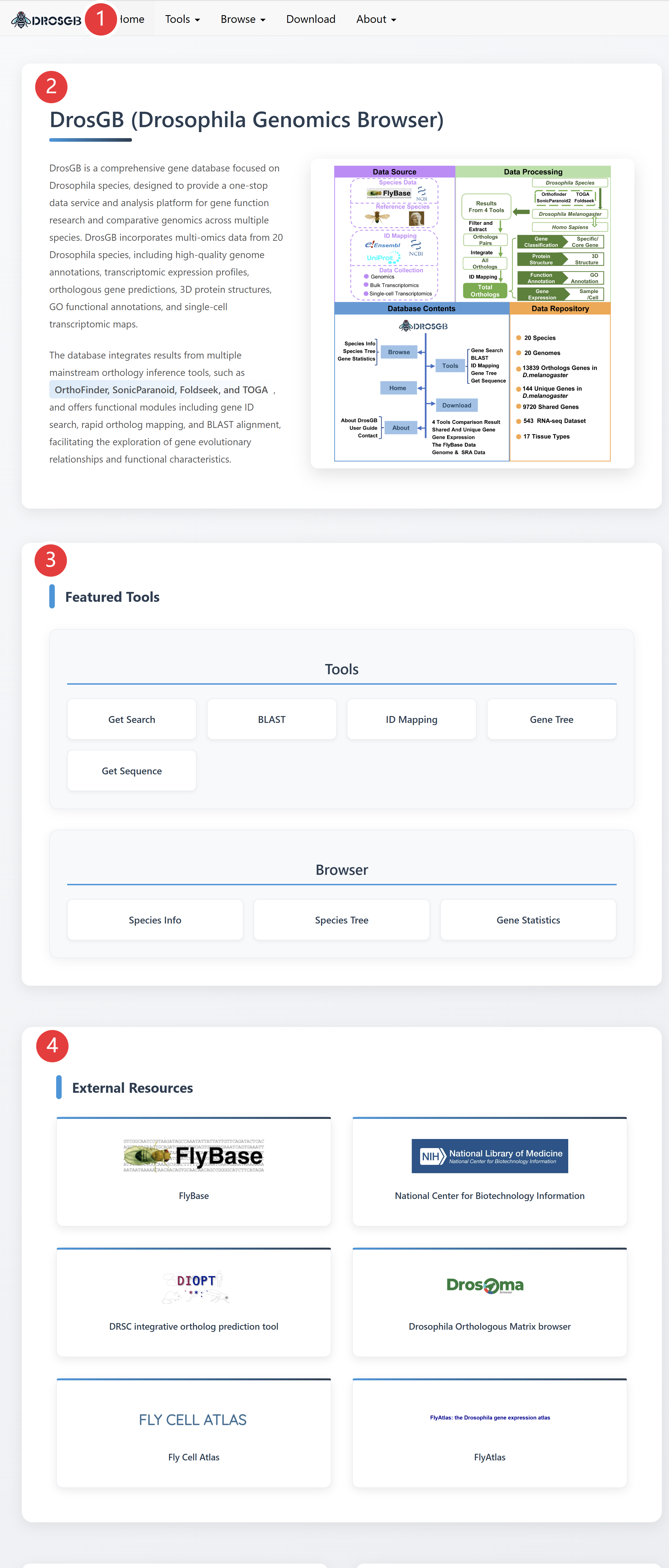
DrosGB is a comprehensive gene database focused on Drosophila species, designed to provide a one-stop data service and analysis platform for gene function research and comparative genomics across multiple species. DrosGB incorporates multi-omics data from 20 Drosophila species, including high-quality genome annotations, transcriptomic expression profiles, orthologous gene predictions, 3D protein structures, and GO functional annotations.
The database integrates results from multiple mainstream orthology inference tools, such as OrthoFinder, SonicParanoid, Foldseek, and TOGA, and offers functional modules including gene ID search, rapid ortholog ID mapping, BLAST alignment, gene tree, and get sequence, facilitating the exploration of gene evolutionary relationships and functional characteristics.
DrosGB is jointly developed and continuously maintained by the research groups of Mo Liu at Guangzhou Medical University and Xiangrui Cai at Nankai University.
1. Homepage
① The top navigation menu contains links to different modules.
② Brief introduction about DrosGB
③ The links of featured tools in the database.
④ External links of related popular websites.
2. Tools
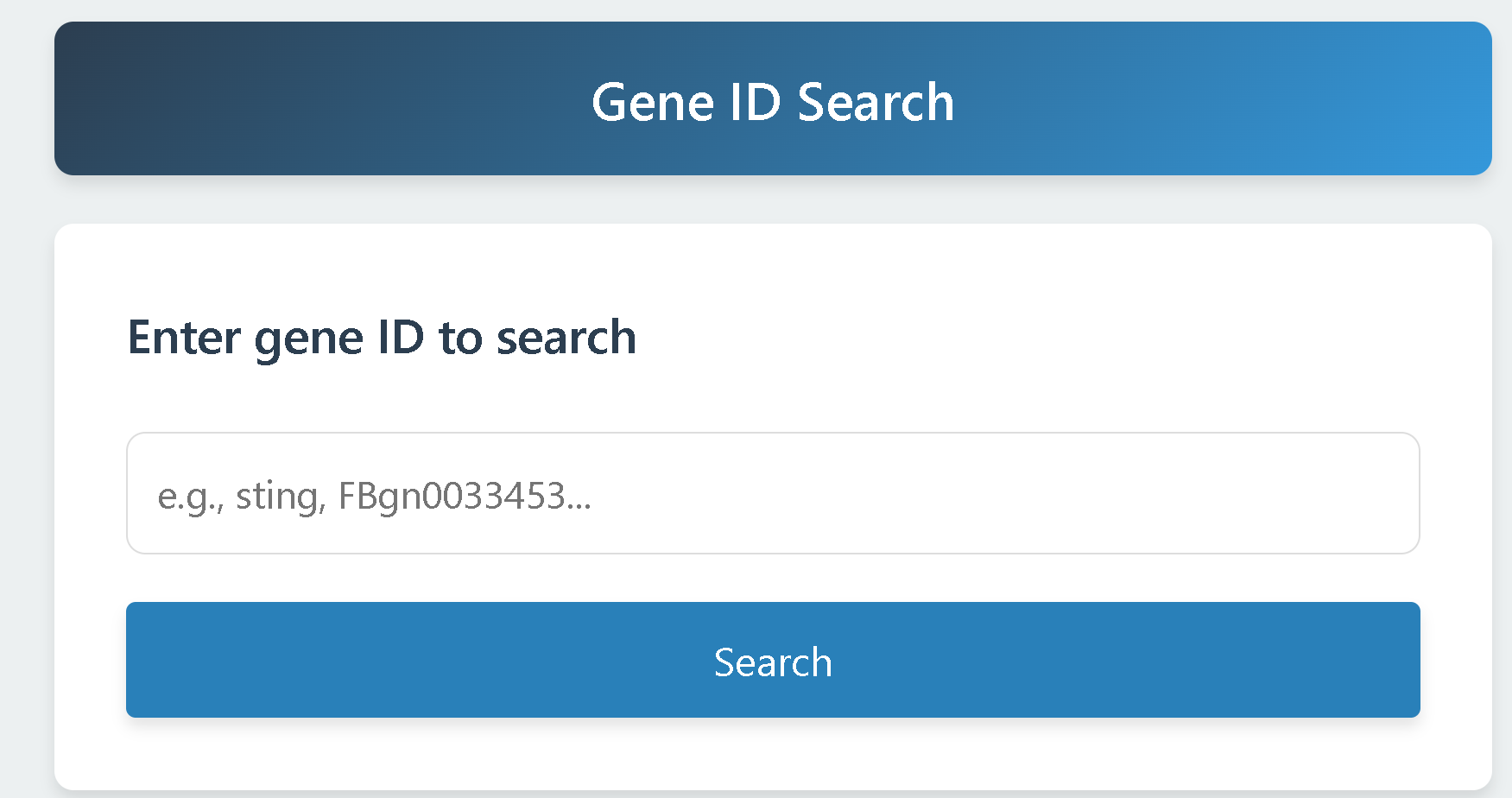
2.1 Gene Search
Users can enter any Gene ID, gene name, or Entrez Gene ID to retrieve the corresponding gene information.
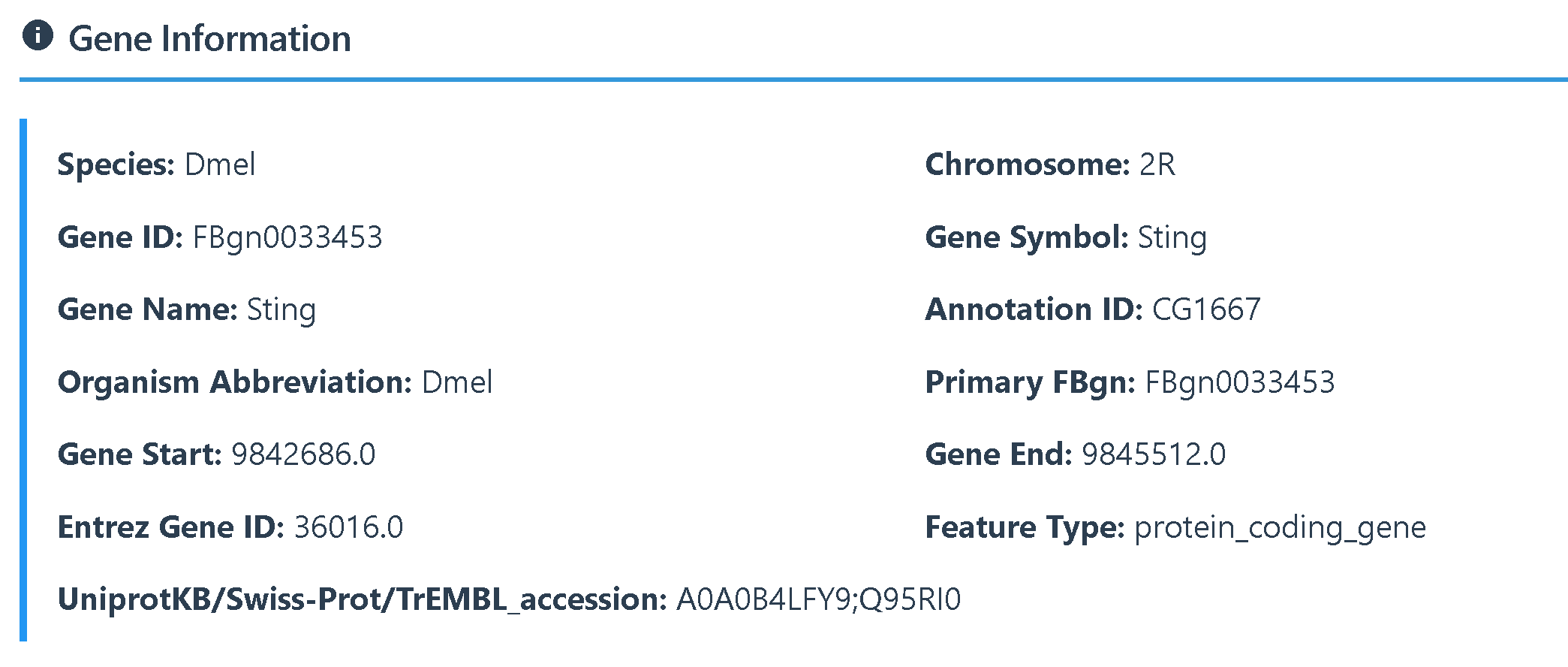
 The results of the gene search can be categorized into Gene Information, Gene Sequences, 3D protein Structure, Gene Expression, Orthologs Results, and Functional Annotation, using the STING gene as an example.
The results of the gene search can be categorized into Gene Information, Gene Sequences, 3D protein Structure, Gene Expression, Orthologs Results, and Functional Annotation, using the STING gene as an example.
(1) Gene Information provides details including the gene ID, gene name, chromosomal location, and aliases used in other databases.
(2) Gene Sequences displays the gene’s full sequence information, including its mRNA, CDS, and protein sequences.

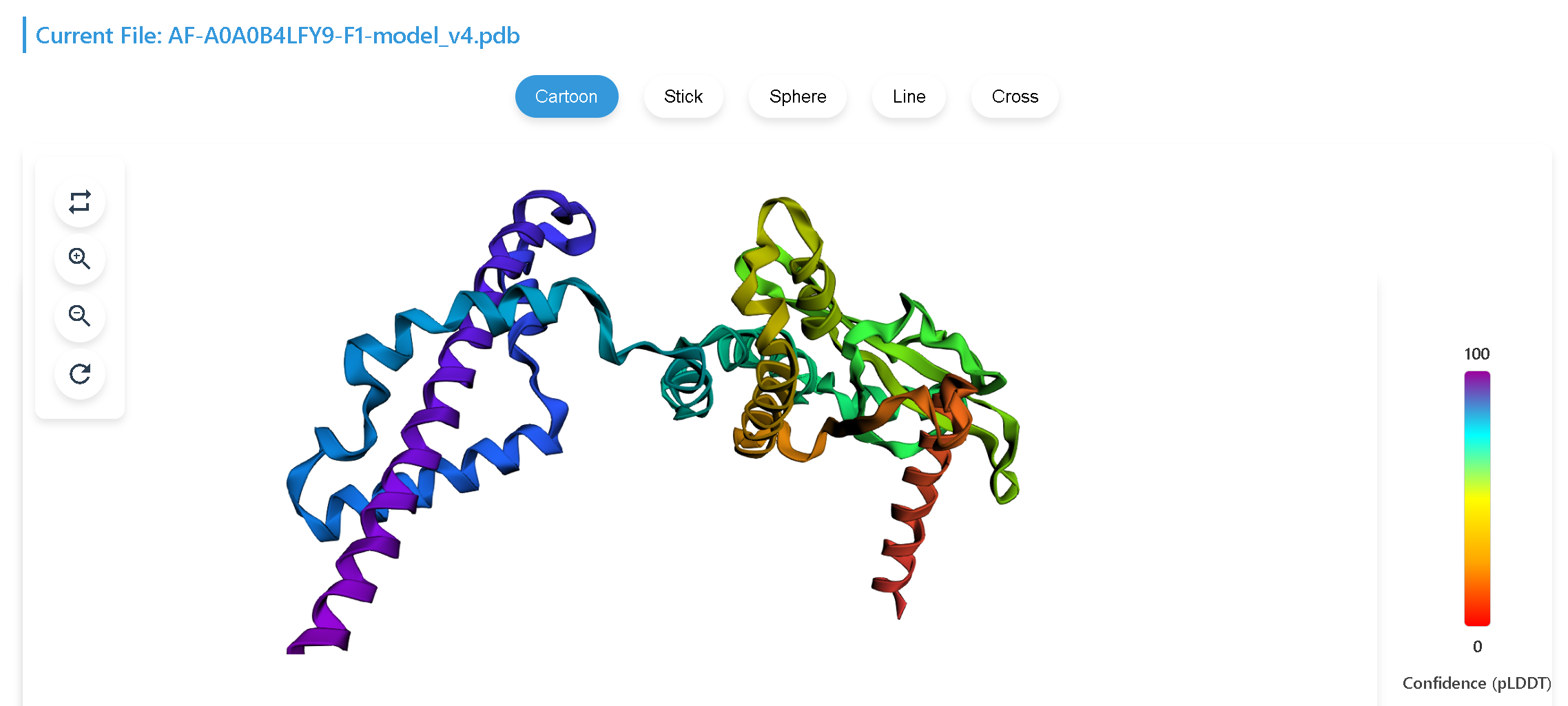
(3) 3D Protein Structure provides the three-dimensional protein structure of the gene.The structure for Drosophila melanogaster is obtained from the AlphaFold database, while those of other Drosophila species are generated using ColaFold.
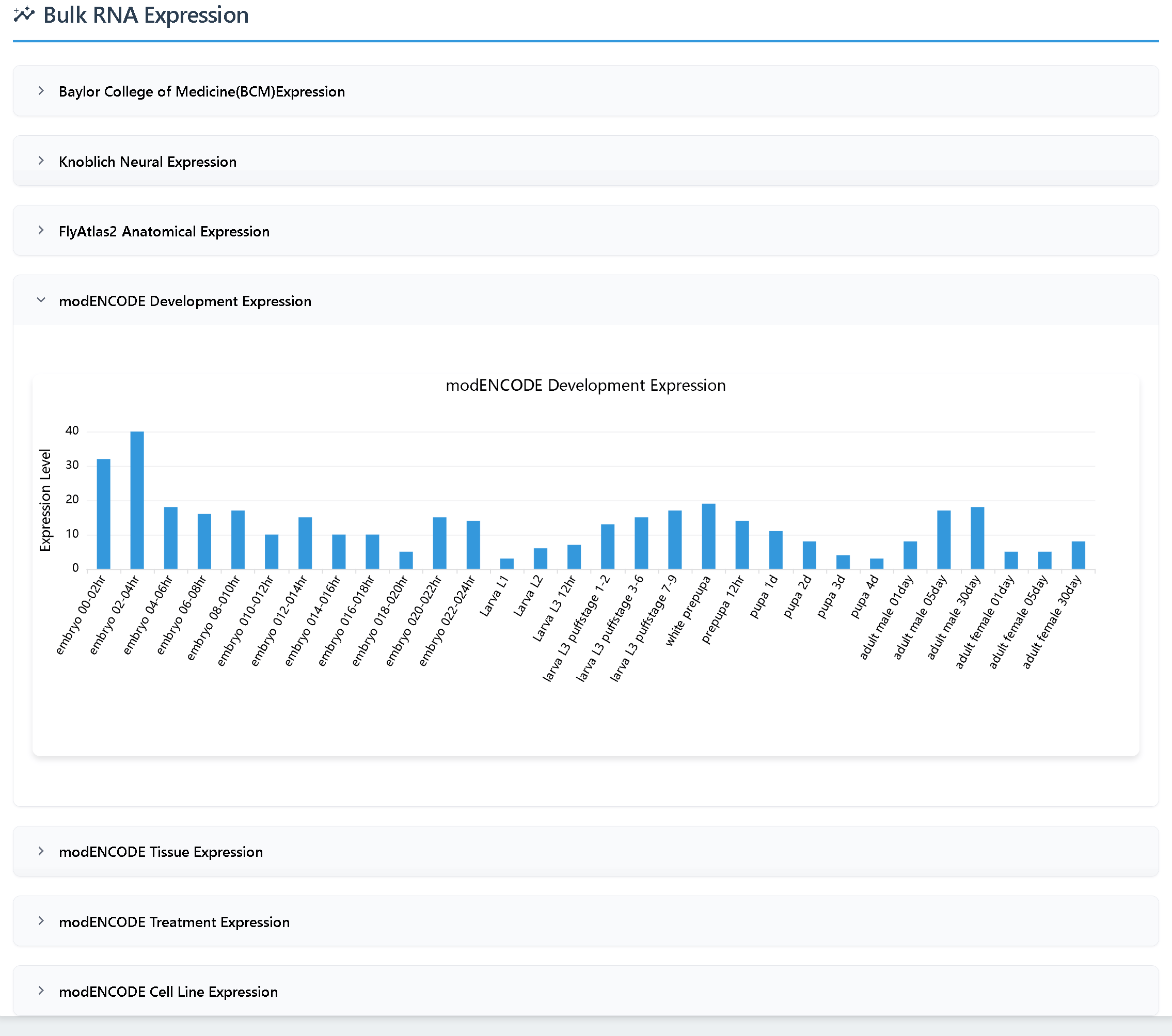
(4)Gene Expression includes two types of data for Drosophila melanogaster:
① Bulk RNA-seq data from seven datasets from FlyBase (RPKM values)
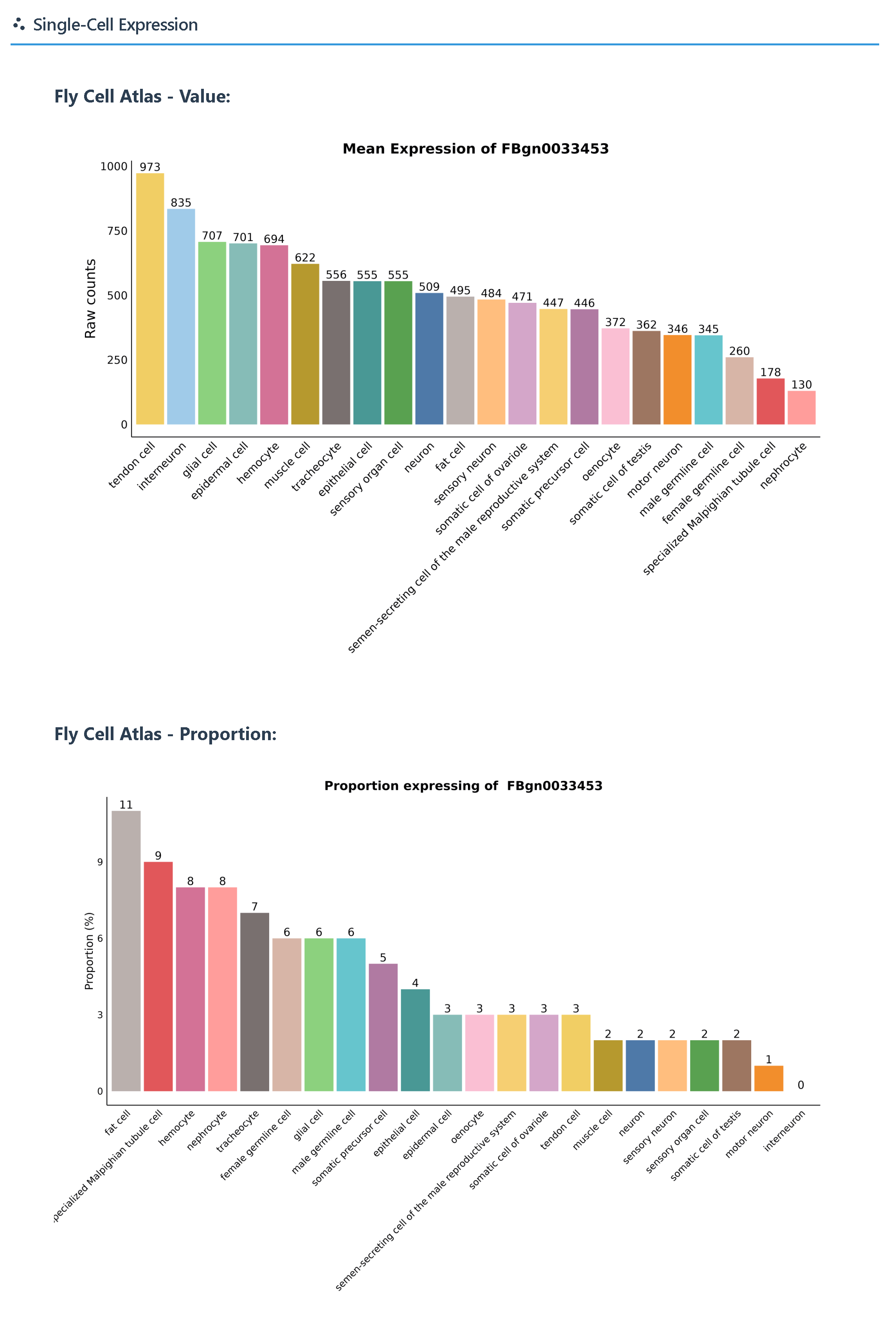
 ② Single-cell expression data from the Fly Cell Atlas (raw counts and proportion), downloaded from FlyBase.
② Single-cell expression data from the Fly Cell Atlas (raw counts and proportion), downloaded from FlyBase.
 For other Drosophila species, only bulk RNA-seq data are available, and their sources are listed in the Download section (SRA data).
For other Drosophila species, only bulk RNA-seq data are available, and their sources are listed in the Download section (SRA data).
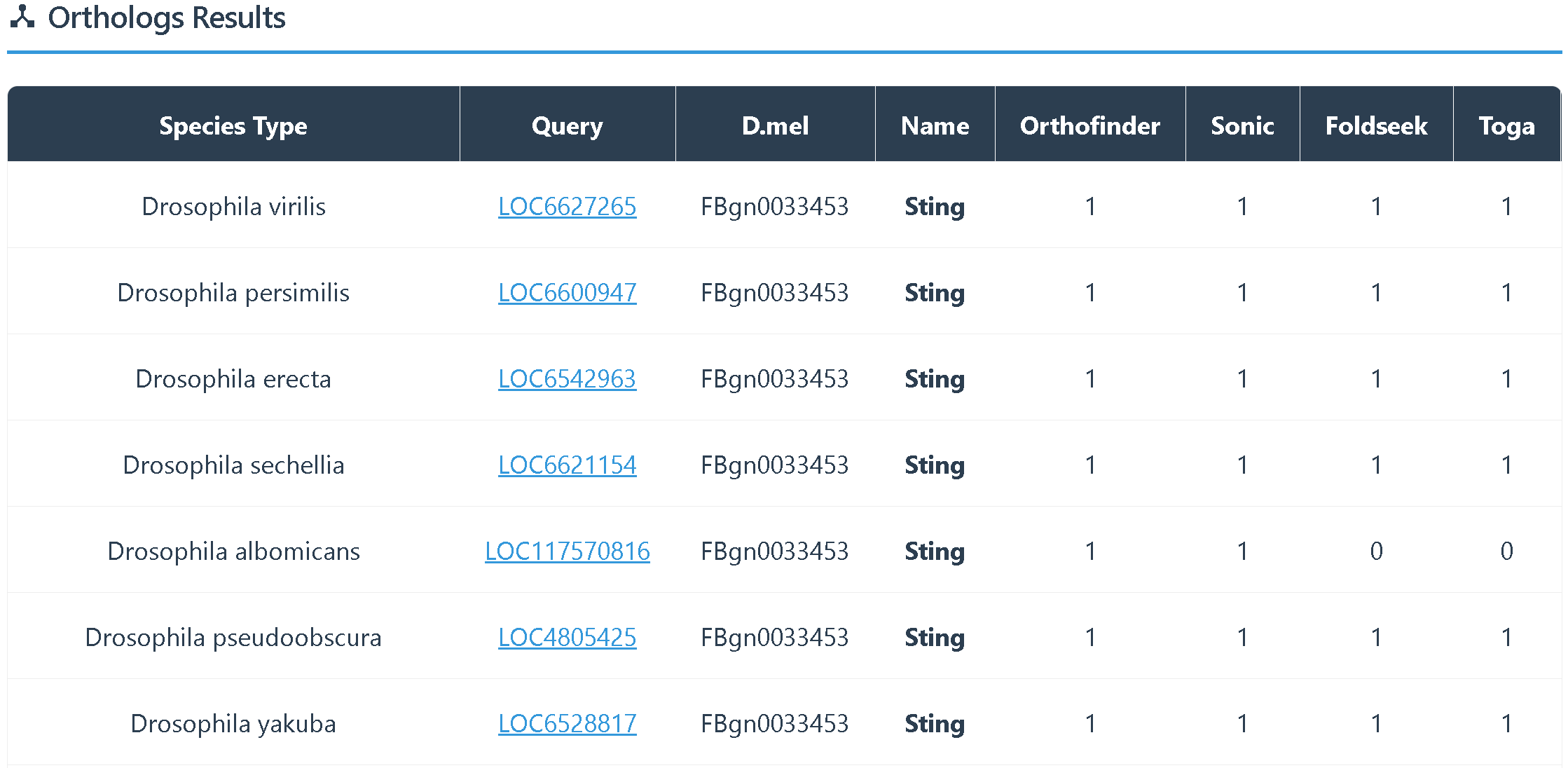
(5)Orthologs Results provides two types of information:
① orthologous genes among other Drosophila species.
 ② orthologous genes in humans.
② orthologous genes in humans.
(6)Functional Annotation provides two types of information:
① basic function
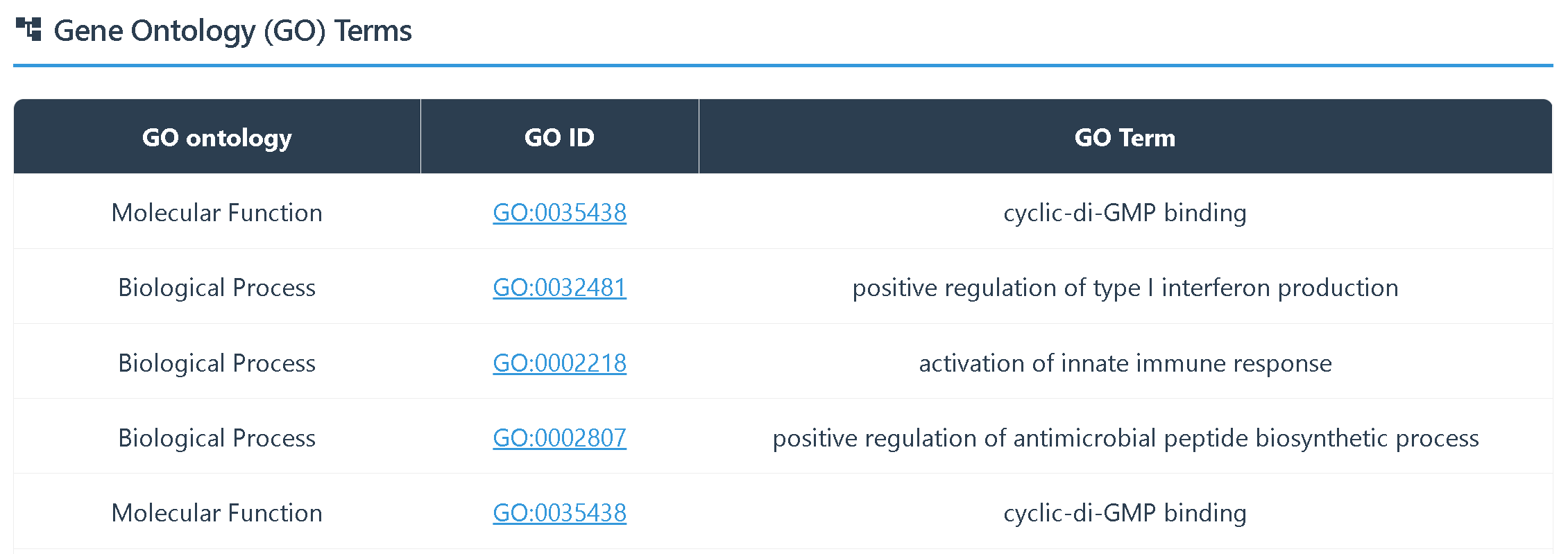
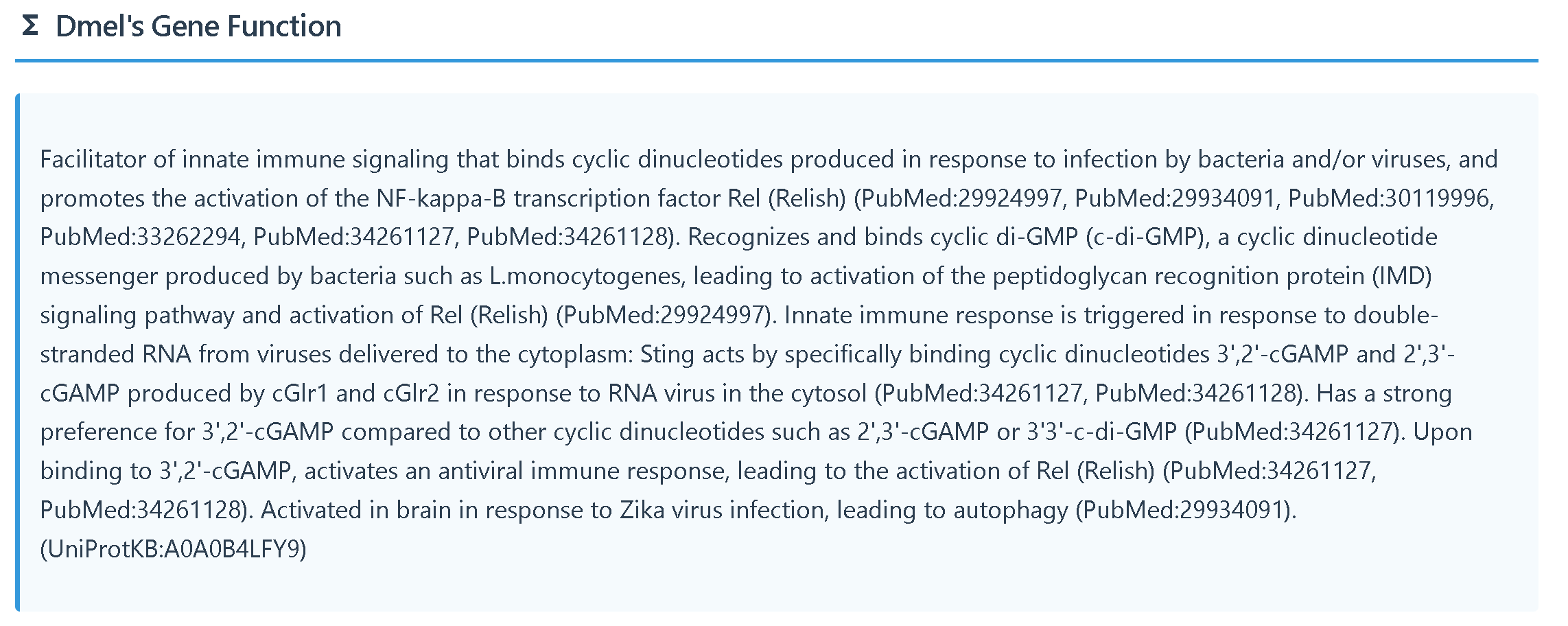 ② GO annotation
② GO annotation
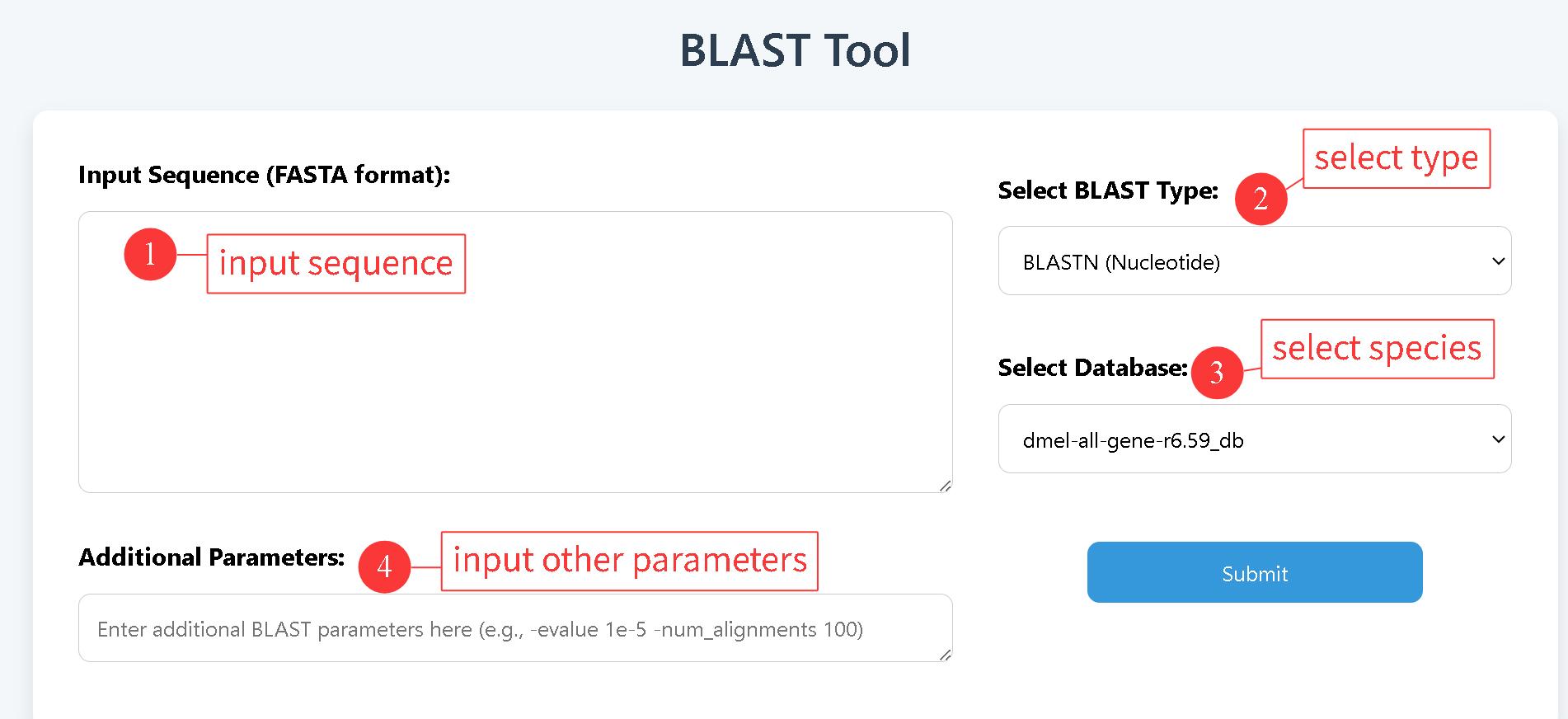
2.2 BLAST
Users can submit query sequences and select either the BLASTN (nucleotide) or BLASTP (protein) model to perform a fast sequence alignment against any of the 20 Drosophila species, enabling the exploration of sequence similarity.
 The BLAST results are presented in the standard outfmt 7 format.
The BLAST results are presented in the standard outfmt 7 format.

2.3 ID Mapping
ID Mapping allows users to input a gene ID and quickly obtain the corresponding orthologous gene IDs in all Drosophila species.
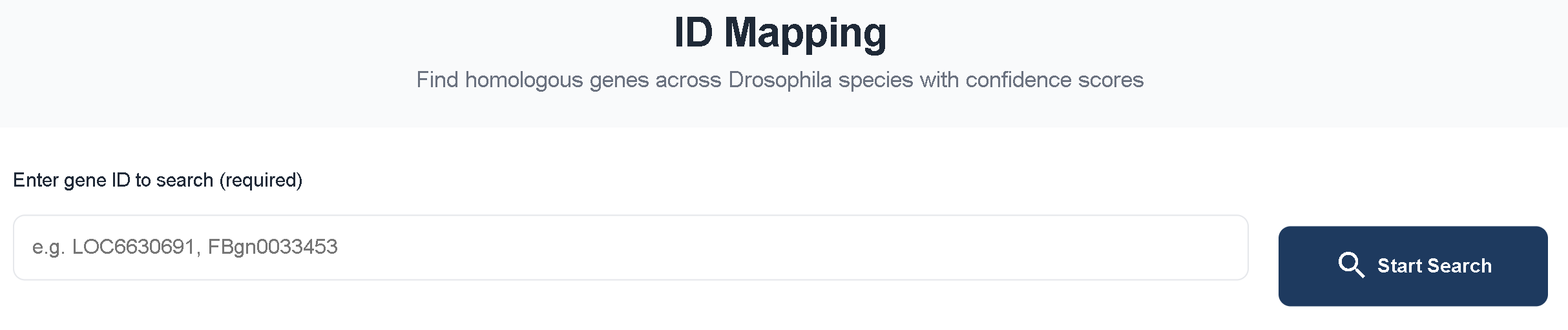 Using FBgn0033453 as an example, the ID Mapping results are categorized into several confidence levels: Confidence Homologous Genes (Sum ≥ 3), Sum = 4, Sum = 3, Sum = 2, and Sum = 1.
Using FBgn0033453 as an example, the ID Mapping results are categorized into several confidence levels: Confidence Homologous Genes (Sum ≥ 3), Sum = 4, Sum = 3, Sum = 2, and Sum = 1.
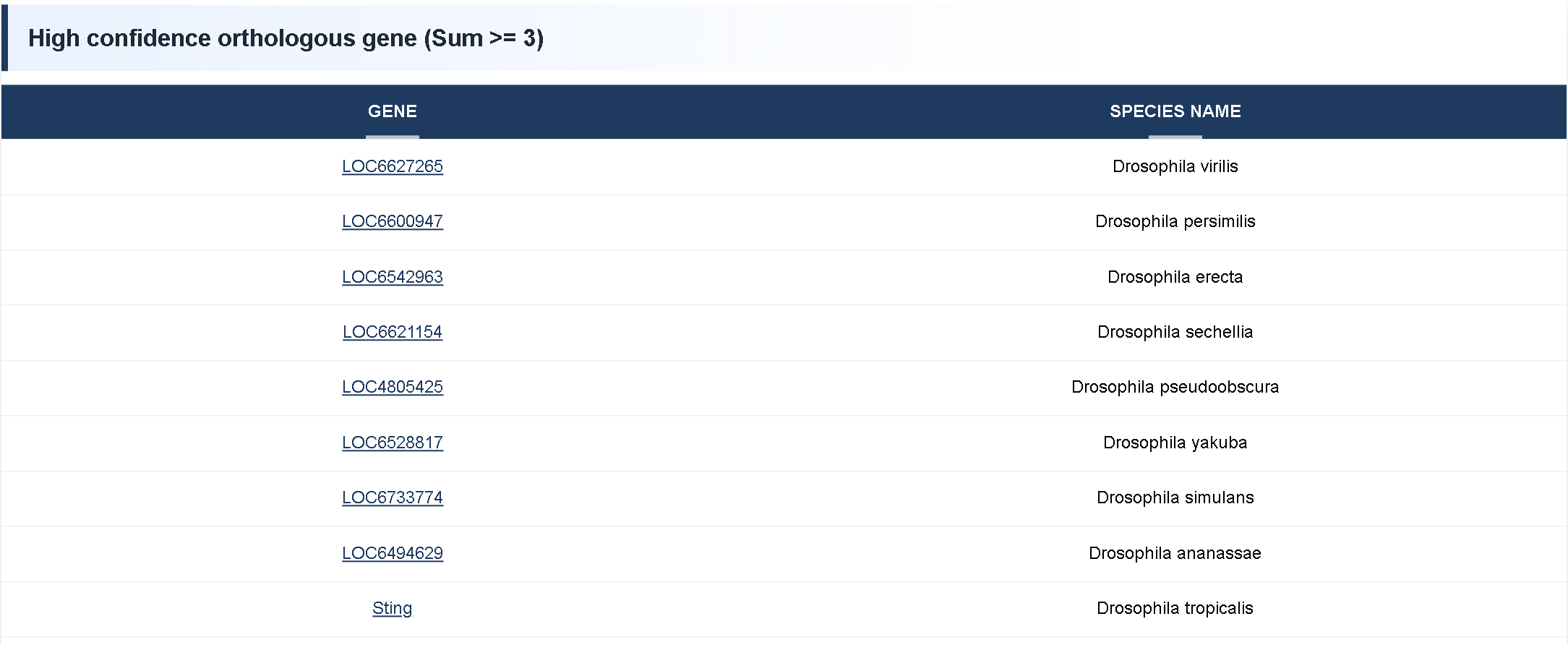
① High Confidence Homologous Genes (Sum ≥ 3) represent orthologous genes supported by at least three orthology detection tools, indicating a high level of reliability. The first column lists the gene ID, and the second column shows the corresponding species.
 ② Sum = 4 includes orthologous genes identified consistently by all four orthology detection tools.
② Sum = 4 includes orthologous genes identified consistently by all four orthology detection tools.Sum = 3 includes genes supported by three orthology detection tools.
Sum = 2 includes genes supported by two orthology detection tools.
Sum = 1 includes genes supported by only one orthology detection tool.
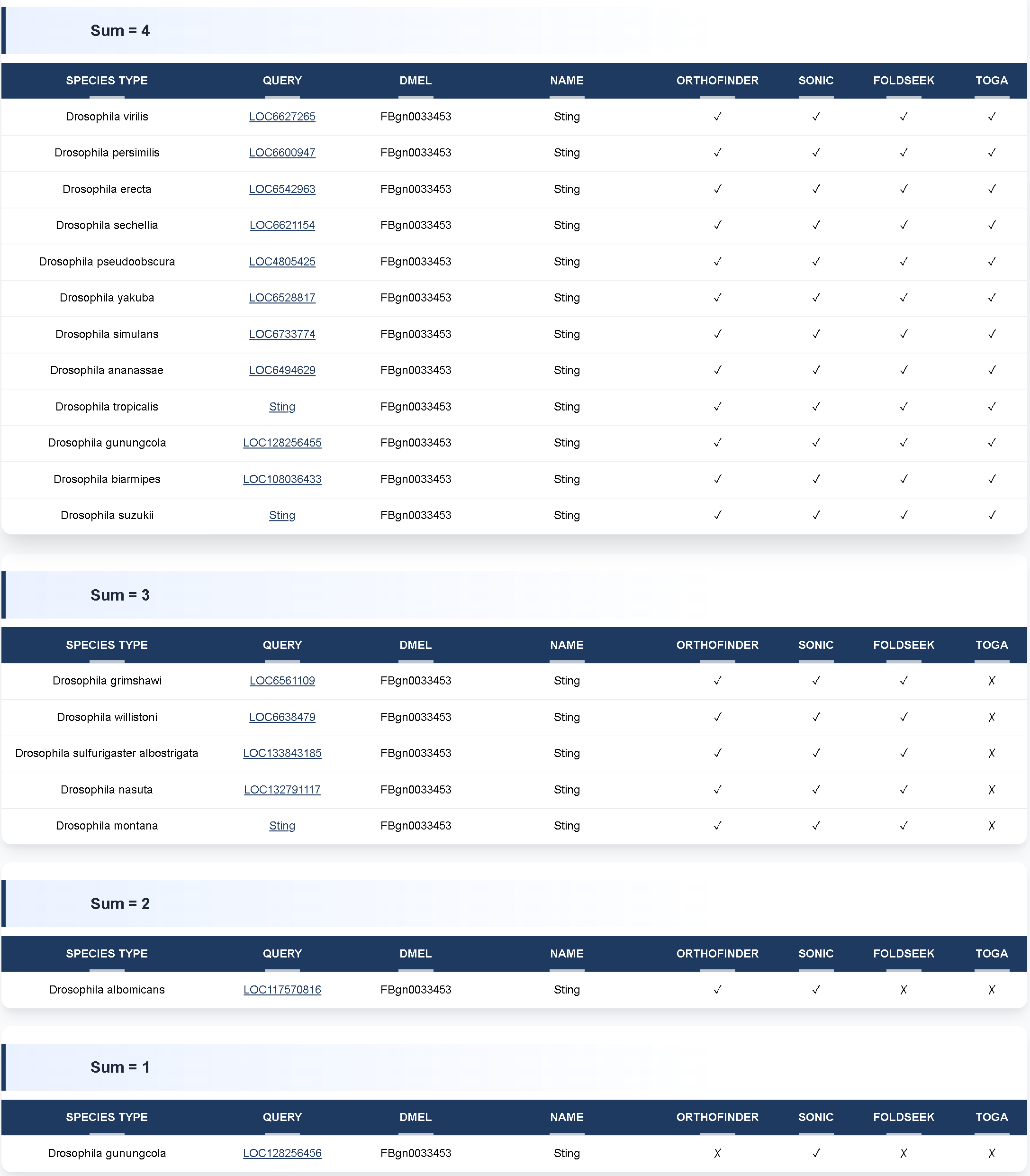 The result table is organized as follows:
The result table is organized as follows:• Column 1: Species name
• Column 2: Orthologous gene ID
• Column 3: Corresponding Drosophila melanogaster gene ID
• Column 4: Gene name
• Columns 5–8: Orthology detection tools used for comparison. ✔ indicates that the tool supports the orthologous relationship, while ✖ indicates that it does not.
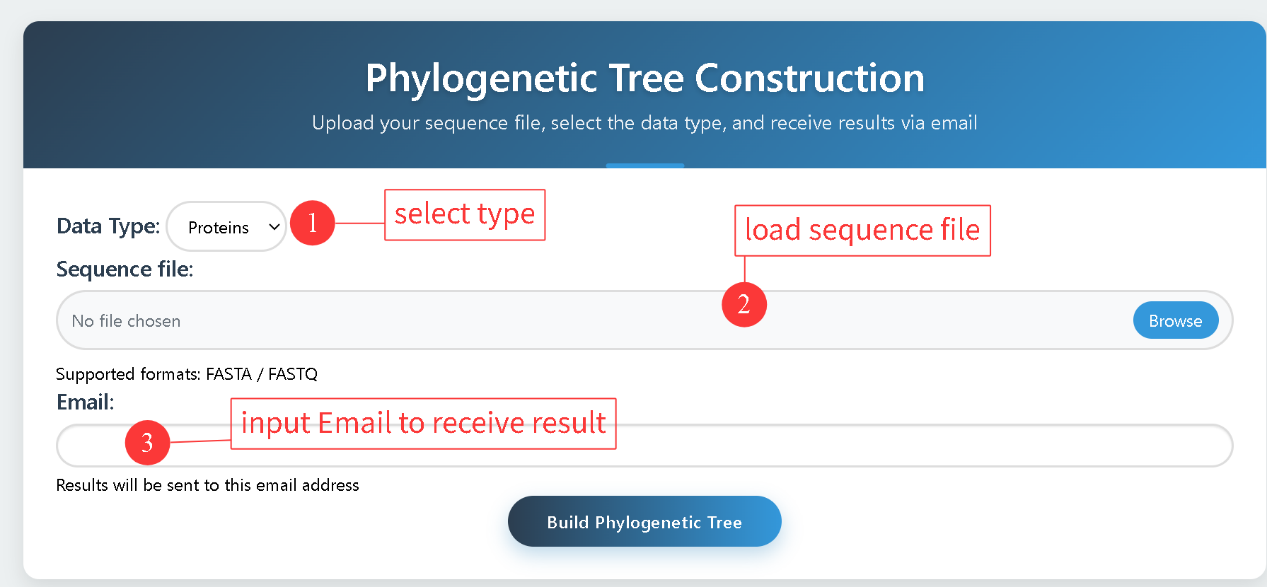
2.4 Gene Tree
Gene Tree allows users to upload a multiple sequence file and choose either protein or CDS sequences for phylogenetic analysis. Sequence alignment is performed using MUSCLE, and the phylogenetic tree is constructed with FastTree. The results are delivered to the user via email.
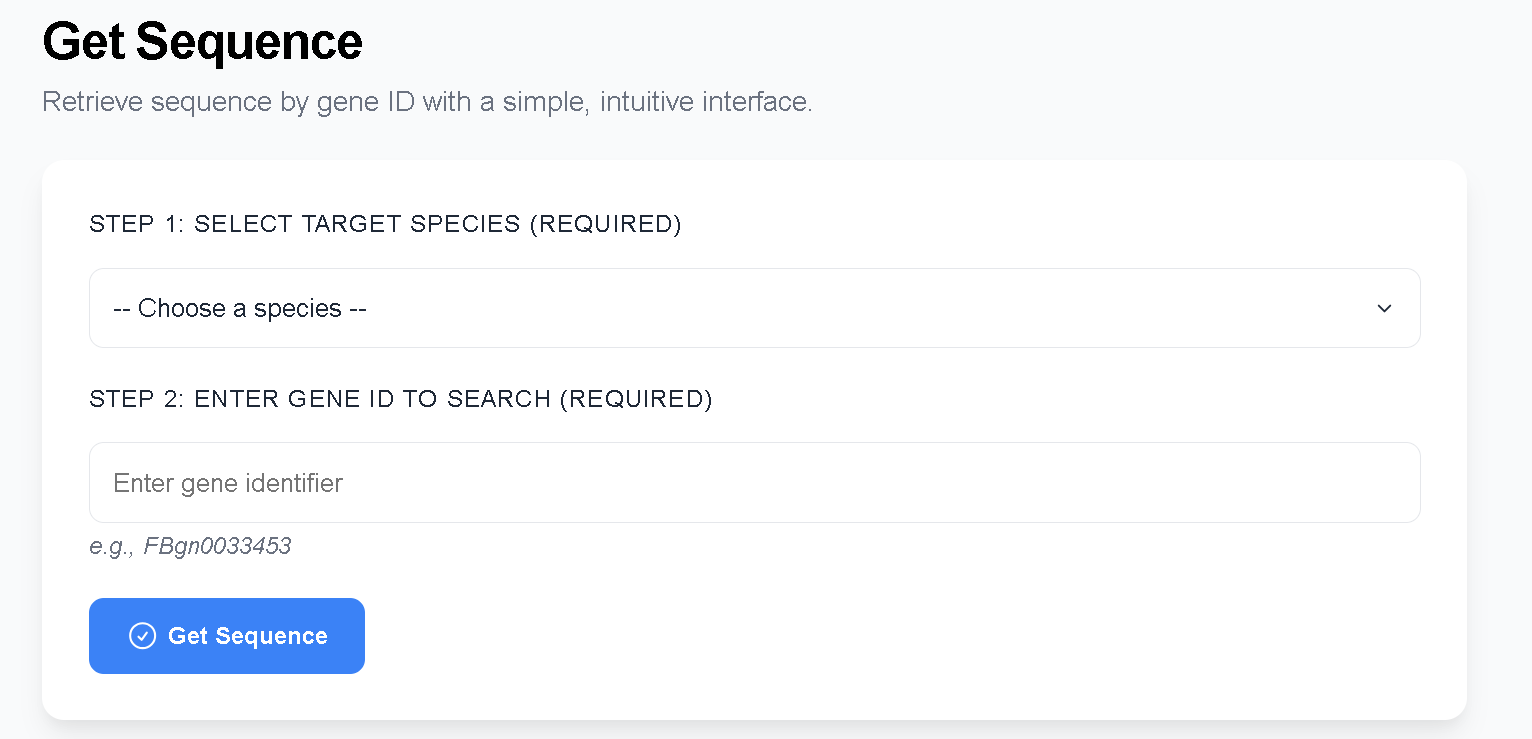
2.5 Get Sequence
Get Sequence allows users to select a species and input a gene ID to obtain the corresponding gene, mRNA, CDS, and protein sequences.
3. Browse
3.1 Species Info
The Species Info module presents detailed introductions to 20 Drosophila species, including their genomic data, annotation information, and relevant literature.

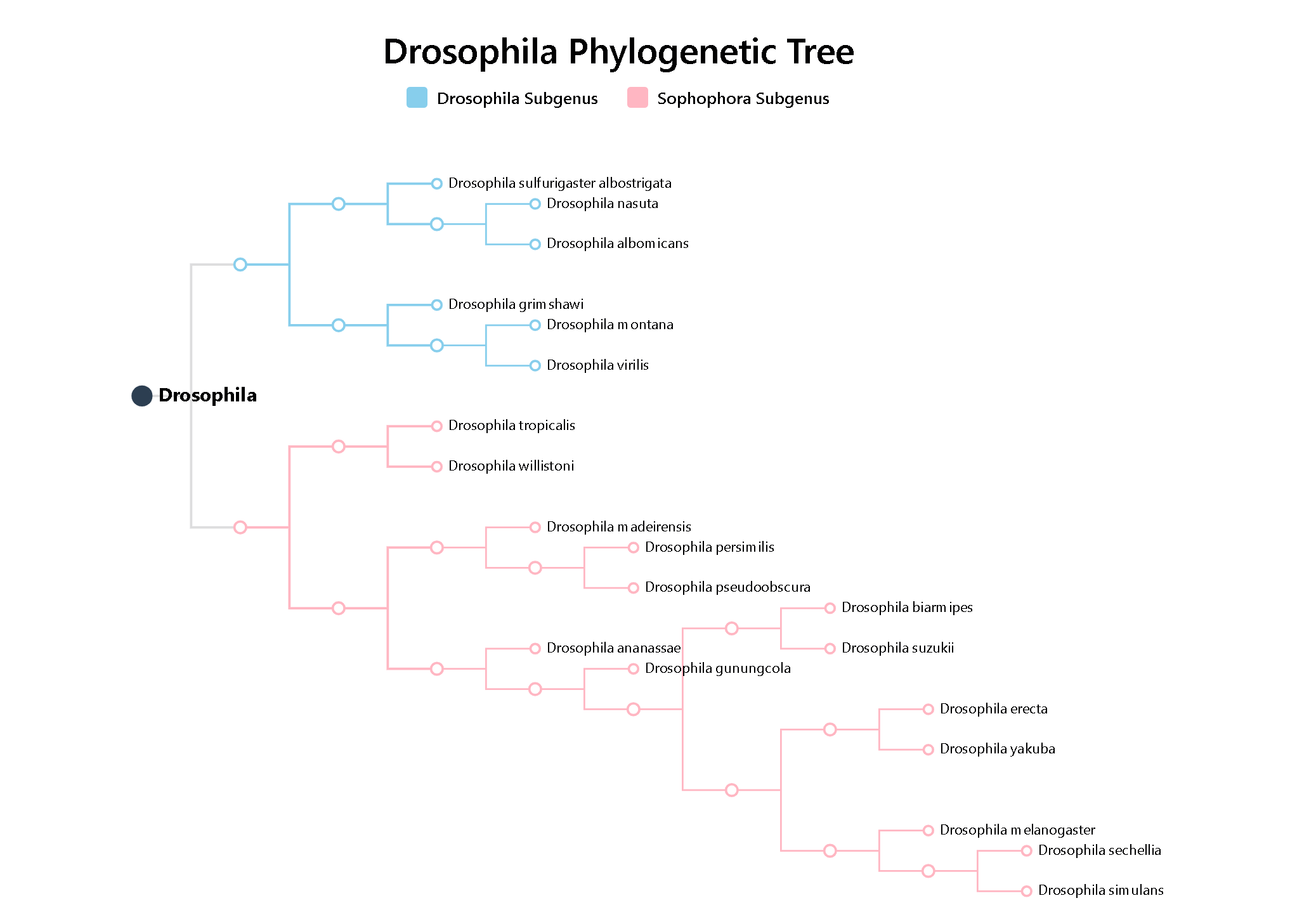
3.2 Species Tree
Species Tree displays the evolutionary relationships among 20 Drosophila species, based on single-copy genes detected by OrthoFinder and analyzed with IQ-TREE. Clicking on a species opens its Species Info page.
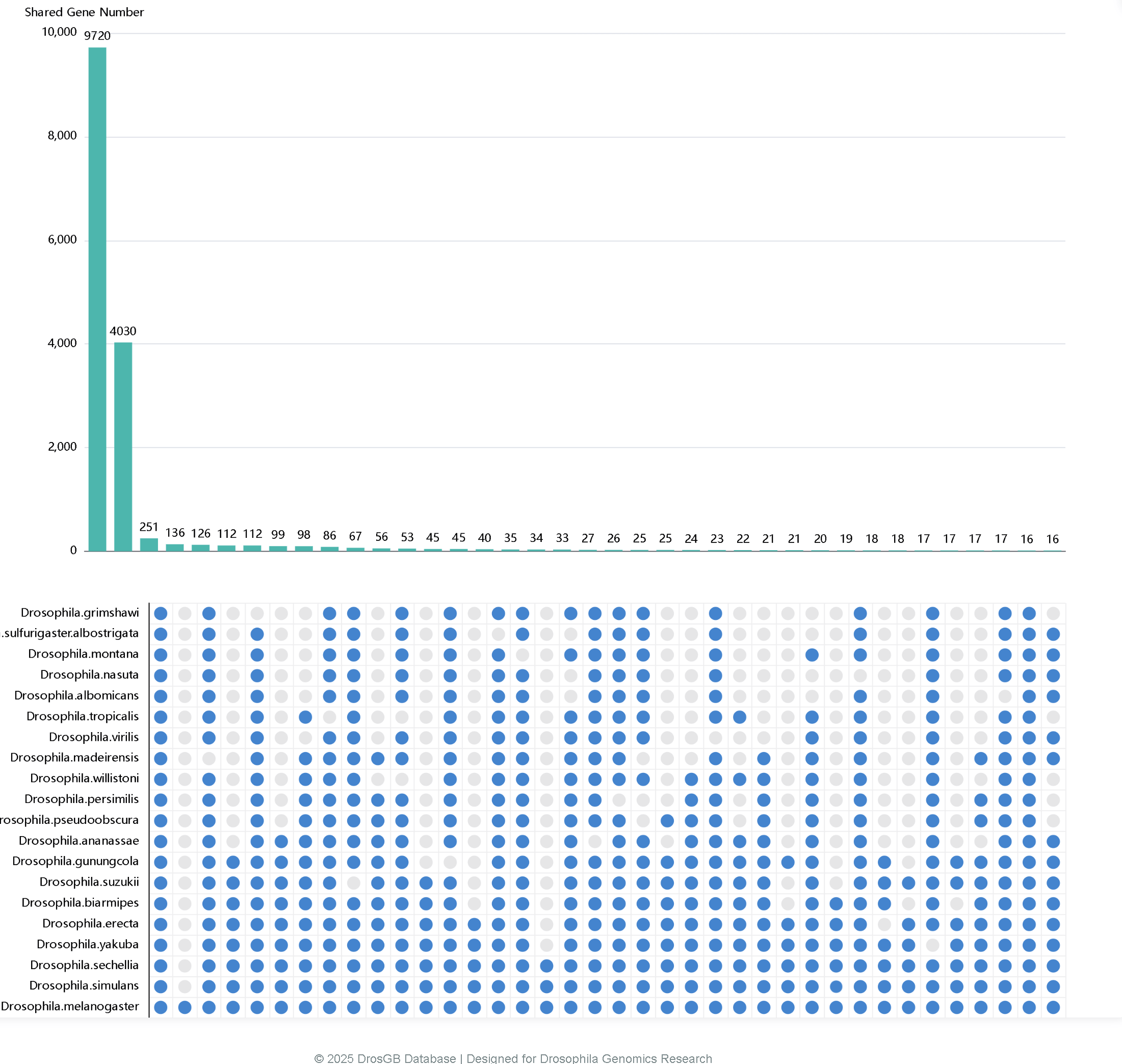
3.3 Gene Statistics
Gene Statistics displays the shared genes among Drosophila species using an UpSet plot. The numbers on the bars indicate the count of shared genes, and users can click on a bar to directly download the corresponding gene list.
4. Download
Download provides access to multiple types of data:
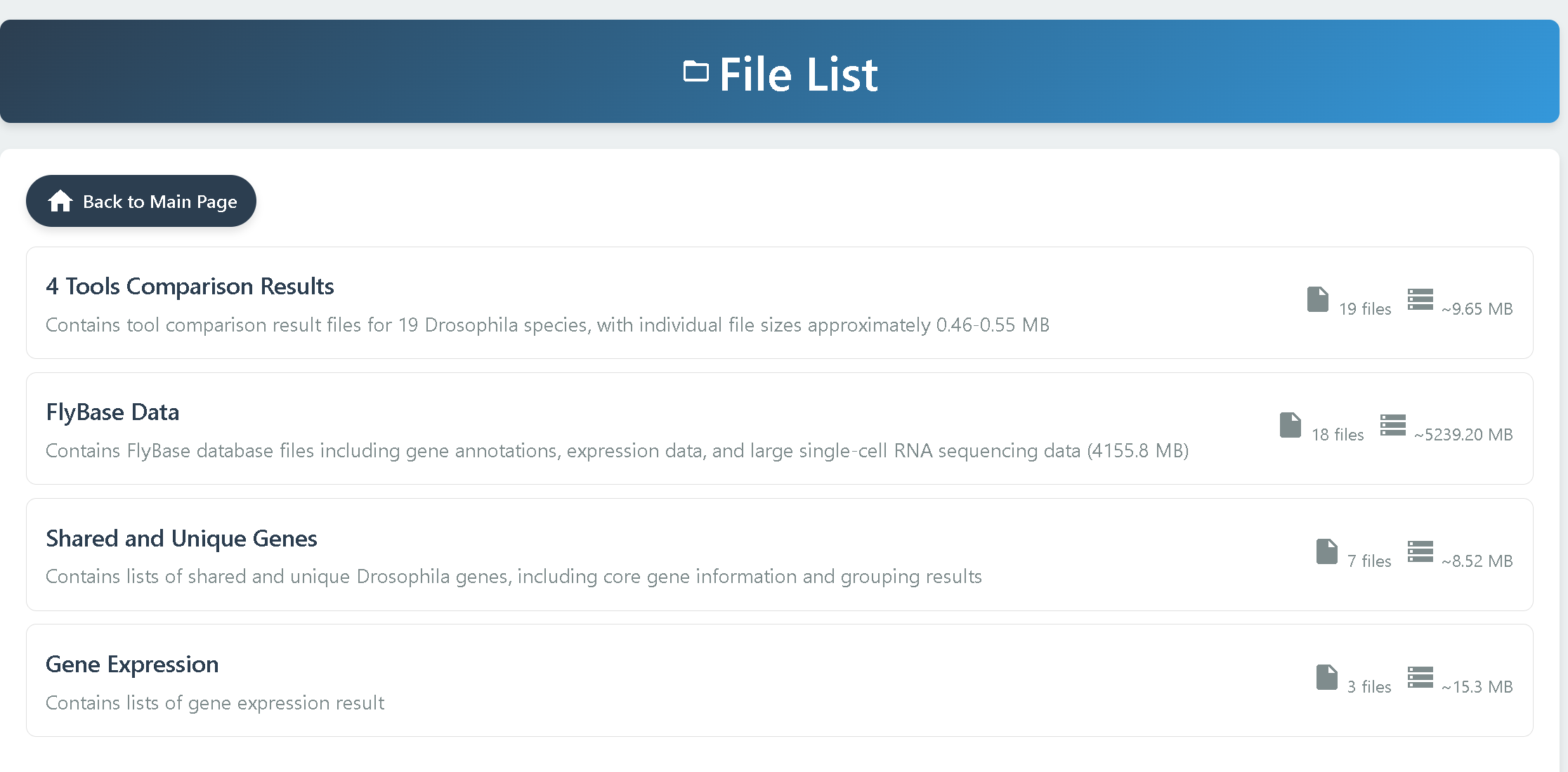
• 4 Tools Comparison Results — Contains ortholog identification results from four tools across 19 Drosophila species, with one file provided for each species.
• FlyBase Data — Provides reference data obtained from the FlyBase database. Detailed descriptions of the files are available in the readme.txt file within the folder.
• Shared and Unique Genes — Includes gene presence and absence information for 20 Drosophila species. File details are described in the readme.txt file within the folder.
• Gene Expression — Contains gene expression datasets. For detailed descriptions, please refer to the readme.txt file within the folder.